Molecular graph
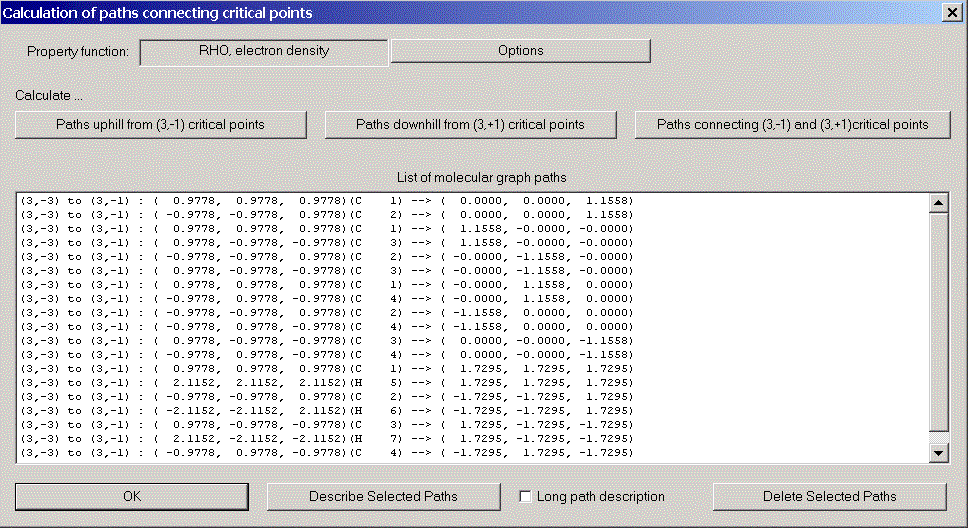
The molecular graph of a molecule for a certain density function consists of all
gradient paths connecting critical points. Usually these paths are calculated for the
charge density.
Of course, all critical points should be known if possible! The program will post a
message if it suspects the existence of a critical point which is not yet known.
The gradient paths of a molecular graph belong to three different categories:
- Paths uphill from (3, -1)-critical points:
The Hessian matrix at a (3, -1)-critical point has one positive eigenvalue. There are two
gradient paths starting at the critical point and going uphill in the direction of the
corresponding eigenvector (positive and negative direction).
When computed for the charge density, these paths form a so-called bond-path which
connects two atoms. The linear combinations of the other two eigenvectors define the
starting directions of downhill gradient paths which constitute an interatomic surface.
The program recognizes interatomic surfaces after the corresponding (3, -1)-critical
points and their bond-paths are calculated.
Bond-paths are also needed for the integration
over atomic basins.
- Paths downhill from (3, +1)-critical points:
The Hessian matrix at a (3, +1)-critical point has one negative eigenvalue. There are two
gradient paths starting at the critical point and going downhill in the direction of the
corresponding eigenvector (positive and negative direction). These paths either terminate at
infinity or a (3, +3)-critical point.
- Paths connecting (3, -1)- and (3, +1)-critical
points: Unfortunately there is not a unique direction for such a gradient
path
(uphill or downhill), since (3, -1)- critical points have two eigenvectors pointing
downhill while (3, +1)-critical points have two eigenvectors pointing uphill.
In order to compute these paths, a bisection iteration procedure has been implemented
which calculates a gradient path in each iteration. The procedure will only work if all
(3, -1)-critical points connected to the (3, +1)-critical point are known. Otherwise a
message is posted. If the iteration fails, you may try to increase the number of bisection
iterations in the Options-dialog. In case of
failure the last computed path is added to the list of molecular graph
paths.
For all computed gradient paths, the program will present a record. Successfully
computed gradient paths will be entered into the right hand side list.
The Options-button brings you to a dialog where you can
manipulate the values used to solve the differential equations of the gradient paths.
- Describe selected gradient paths displays a short or long
description of the highlighted gradient paths.
-
Delete selected gradient paths removes the highlighted gradient paths
from the list.
-
OK closes the dialog.
